Quantify dataset specification#
See also
The complete source code of this tutorial can be found in
Show code cell content
from pathlib import Path
import matplotlib.pyplot as plt
import xarray as xr
from rich import pretty
import quantify_core.data.dataset_adapters as dadapters
import quantify_core.data.dataset_attrs as dattrs
from quantify_core.data import handling as dh
from quantify_core.utilities import dataset_examples
from quantify_core.utilities.examples_support import round_trip_dataset
from quantify_core.utilities.inspect_utils import display_source_code
pretty.install()
dh.set_datadir(Path.home() / "quantify-data") # change me!
This document describes the Quantify dataset specification.
Here we focus on the concepts and terminology specific to the Quantify dataset.
It is based on the Xarray dataset, hence, we assume basic familiarity with the xarray.Dataset.
If you are not familiar with it, we highly recommend to first have a look at our Xarray - brief introduction for a brief overview.
Coordinates and Variables#
The Quantify dataset is an xarray dataset that follows certain conventions. We define “subtypes” of xarray coordinates and variables:
Main coordinate(s)#
Xarray Coordinates that have an attribute
is_main_coordset toTrue.Often correspond to physical coordinates, e.g., a signal frequency or amplitude.
Often correspond to quantities set through
Settables.The dataset must have at least one main coordinate.
Example: In some cases, the idea of a coordinate does not apply, however a main coordinate in the dataset is required. A simple “index” coordinate should be used, e.g., an array of integers.
See also the method
get_main_coords().
Secondary coordinate(s)#
A ubiquitous example is the coordinates that are used by “calibration” points.
Similar to main coordinates, but intended to serve as the coordinates of secondary variables.
Xarray Coordinates that have an attribute
is_main_coordset toFalse.See also
get_secondary_coords().
Main variable(s)#
Xarray Variables that have an attribute
is_main_varset toTrue.Often correspond to a physical quantity being measured, e.g., the signal magnitude at a specific frequency measured on a metal contact of a quantum chip.
Often correspond to quantities returned by
Gettables.See also
get_main_vars().
Secondary variables(s)#
Again, the ubiquitous example is “calibration” datapoints.
Similar to main variables, but intended to serve as reference data for other main variables (e.g., calibration data).
Xarray Variables that have an attribute
is_main_varset toFalse.The “assignment” of secondary variables to main variables should be done using
relationships.See also
get_secondary_vars().
Note
In this document we show exemplary datasets to highlight the details of the Quantify dataset specification. However, for completeness, we always show a valid Quantify dataset with all the required properties.
In order to follow the rest of this specification more easily have a look at the example below. It should give you a more concrete feeling of the details that are exposed afterward. See Quantify dataset - examples for an exemplary dataset.
We use the
mk_two_qubit_chevron_dataset() to
generate our dataset.
Source code for generating the dataset below
display_source_code(dataset_examples.mk_two_qubit_chevron_dataset)
def mk_two_qubit_chevron_dataset(**kwargs) -> xr.Dataset:
"""
Generates a dataset that look similar to a two-qubit Chevron experiment.
Parameters
----------
**kwargs
Keyword arguments passed to :func:`~.mk_two_qubit_chevron_data`.
Returns
-------
:
A mock Quantify dataset.
"""
amp_values, time_values, pop_q0, pop_q1 = mk_two_qubit_chevron_data(**kwargs)
dims_q0 = dims_q1 = ("repetitions", "main_dim")
pop_q0_attrs = mk_main_var_attrs(
long_name="Population Q0", unit="", has_repetitions=True
)
pop_q1_attrs = mk_main_var_attrs(
long_name="Population Q1", unit="", has_repetitions=True
)
data_vars = dict(
pop_q0=(dims_q0, pop_q0, pop_q0_attrs),
pop_q1=(dims_q1, pop_q1, pop_q1_attrs),
)
dims_amp = dims_time = ("main_dim",)
amp_attrs = mk_main_coord_attrs(long_name="Amplitude", unit="V")
time_attrs = mk_main_coord_attrs(long_name="Time", unit="s")
coords = dict(
amp=(dims_amp, amp_values, amp_attrs),
time=(dims_time, time_values, time_attrs),
)
dataset_attrs = mk_dataset_attrs()
dataset = xr.Dataset(data_vars=data_vars, coords=coords, attrs=dataset_attrs)
return dataset
dataset = dataset_examples.mk_two_qubit_chevron_dataset()
assert dataset == round_trip_dataset(dataset) # confirm read/write
2D example#
In the dataset below we have two main coordinates amp and time; and two main
variables pop_q0 and pop_q1.
Both main coordinates “lie” along a single xarray dimension, main_dim.
Both main variables lie along two xarray dimensions main_dim and repetitions.
dataset
<xarray.Dataset> Size: 115kB
Dimensions: (repetitions: 5, main_dim: 1200)
Coordinates:
amp (main_dim) float64 10kB 0.45 0.4534 0.4569 ... 0.5431 0.5466 0.55
time (main_dim) float64 10kB 0.0 0.0 0.0 0.0 ... 1e-07 1e-07 1e-07 1e-07
Dimensions without coordinates: repetitions, main_dim
Data variables:
pop_q0 (repetitions, main_dim) float64 48kB 0.5 0.5 0.5 ... 0.4818 0.5
pop_q1 (repetitions, main_dim) float64 48kB 0.5 0.5 0.5 ... 0.5371 0.5
Attributes:
tuid: 20241014-175532-795-90ad16
dataset_name:
dataset_state: None
timestamp_start: None
timestamp_end: None
quantify_dataset_version: 2.0.0
software_versions: {}
relationships: []
json_serialize_exclude: []Please note how the underlying arrays for the coordinates are structured! Even for “gridded” data, the coordinates are arranged in arrays that match the dimensions of the variables in the xarray. This is done so that the data can support more complex scenarios, such as irregularly spaced samples and measurements taken at unknown locations.
n_points = 110 # only plot a few points for clarity
_, axs = plt.subplots(4, 1, sharex=True, figsize=(10, 10))
dataset.amp[:n_points].plot(
ax=axs[0], marker=".", color="C0", label=dataset.amp.long_name
)
dataset.time[:n_points].plot(
ax=axs[1], marker=".", color="C1", label=dataset.time.long_name
)
_ = dataset.pop_q0.sel(repetitions=0)[:n_points].plot(
ax=axs[2], marker=".", color="C2", label=dataset.pop_q0.long_name
)
_ = dataset.pop_q1.sel(repetitions=0)[:n_points].plot(
ax=axs[3], marker=".", color="C3", label=dataset.pop_q1.long_name
)
for ax in axs:
ax.legend()
ax.grid()
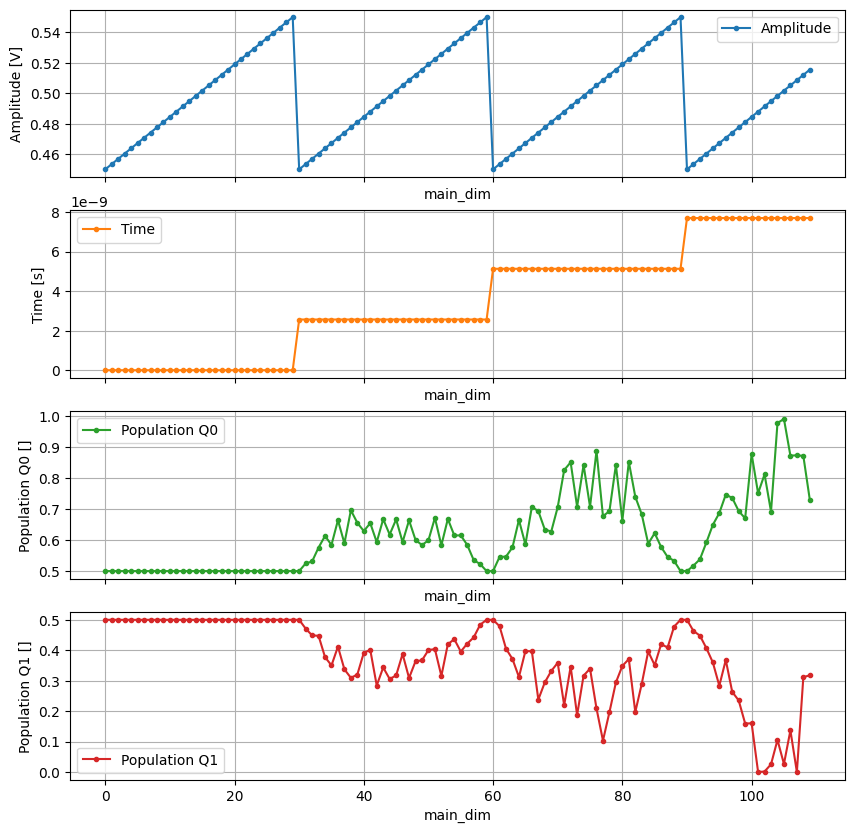
As seen above, in the Quantify dataset the main coordinates do not explicitly index the main variables because not all use-cases fit within this paradigm. However, when possible, the Quantify dataset can be reshaped to take advantage of the xarray built-in utilities.
dataset_gridded = dh.to_gridded_dataset(
dataset,
dimension="main_dim",
coords_names=dattrs.get_main_coords(dataset),
)
dataset_gridded.pop_q0.plot.pcolormesh(x="amp", col="repetitions")
_ = dataset_gridded.pop_q1.plot.pcolormesh(x="amp", col="repetitions")
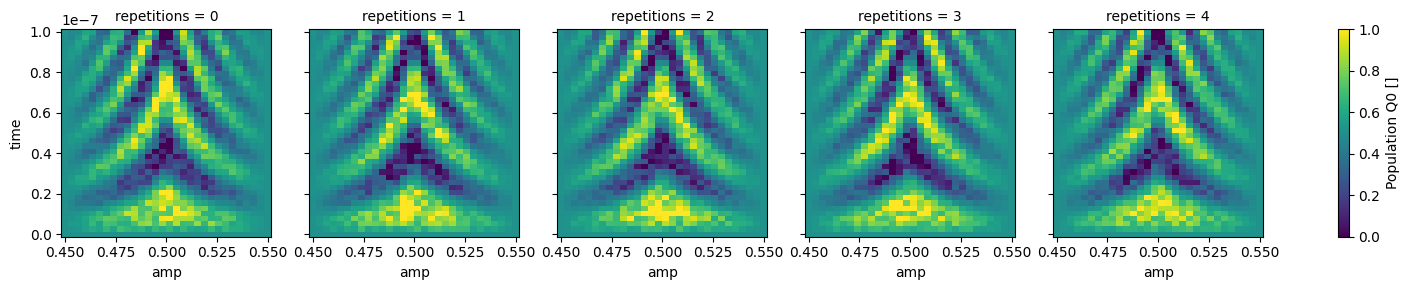
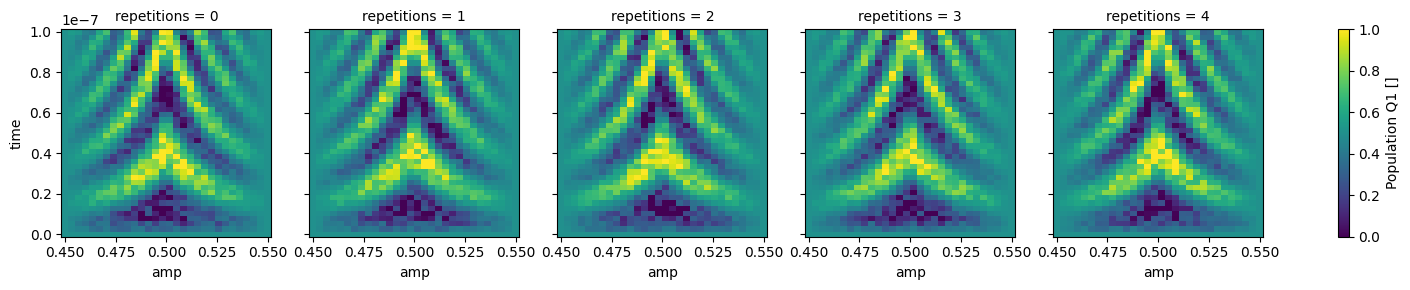
Dimensions#
The main variables and coordinates present in a Quantify dataset have the following required and optional xarray dimensions:
Main dimension(s) [Required]#
The main dimensions comply with the following:
The outermost dimension of any main coordinate/variable, OR the second outermost dimension if the outermost one is a repetitions dimension.
Do not require to be explicitly specified in any metadata attributes, instead utilities for extracting them are provided. See
get_main_dims()which simply applies the rule above while inspecting all the main coordinates and variables present in the dataset.The dataset must have at least one main dimension.
Note on nesting main dimensions
Nesting main dimensions is allowed in principle and such examples are provided but it should be considered an experimental feature.
Intuition: intended primarily for time series, also known as “time trace” or simply trace. See T1 experiment storing digitized signals for all shots for an example.
Secondary dimension(s) [Optional]#
Equivalent to the main dimensions but used by the secondary coordinates and variables. The secondary dimensions comply with the following:
The outermost dimension of any secondary coordinate/variable, OR the second outermost dimension if the outermost one is a repetitions dimension.
Do not require to be explicitly specified in any metadata attributes, instead utilities for extracting them are provided. See
get_secondary_dims()which simply applies the rule above while inspecting all the secondary coordinates and variables present in the dataset.
Repetitions dimension(s) [Optional]#
Repetition dimensions comply with the following:
Any dimension that is the outermost dimension of a main or secondary variable when its attribute
QVarAttrs.has_repetitionsis set toTrue.Intuition for this xarray dimension(s): the equivalent would be to have
dataset_reptition_0.hdf5,dataset_reptition_1.hdf5, etc. where each dataset was obtained from repeating exactly the same experiment. Instead we define an outer dimension for this.Default behavior of (live) plotting and analysis tools can be to average the main variables along the repetitions dimension(s).
Can be the outermost dimension of the main (and secondary) variables.
Variables can lie along one (and only one) repetitions outermost dimension.
Example datasets with repetition#
As shown in the Xarray - brief introduction an xarray dimension can be indexed by a coordinate variable. In this example the repetitions dimension is indexed by the repetitions xarray coordinate. Note that in an xarray dataset, a dimension and a data variable or a coordinate can share the same name. This might be confusing at first. It takes just a bit of dataset manipulation practice to gain an intuition for how it works.
coord_dims = ("repetitions",)
coord_values = ["A", "B", "C", "D", "E"]
dataset_indexed_rep = xr.Dataset(coords=dict(repetitions=(coord_dims, coord_values)))
dataset_indexed_rep
<xarray.Dataset> Size: 20B
Dimensions: (repetitions: 5)
Coordinates:
* repetitions (repetitions) <U1 20B 'A' 'B' 'C' 'D' 'E'
Data variables:
*empty*# merge with the previous dataset
dataset_rep = dataset.merge(dataset_indexed_rep, combine_attrs="drop_conflicts")
assert dataset_rep == round_trip_dataset(dataset_rep) # confirm read/write
dataset_rep
<xarray.Dataset> Size: 115kB
Dimensions: (repetitions: 5, main_dim: 1200)
Coordinates:
amp (main_dim) float64 10kB 0.45 0.4534 0.4569 ... 0.5466 0.55
time (main_dim) float64 10kB 0.0 0.0 0.0 0.0 ... 1e-07 1e-07 1e-07
* repetitions (repetitions) <U1 20B 'A' 'B' 'C' 'D' 'E'
Dimensions without coordinates: main_dim
Data variables:
pop_q0 (repetitions, main_dim) float64 48kB 0.5 0.5 0.5 ... 0.4818 0.5
pop_q1 (repetitions, main_dim) float64 48kB 0.5 0.5 0.5 ... 0.5371 0.5
Attributes:
tuid: 20241014-175532-795-90ad16
dataset_name:
dataset_state: None
timestamp_start: None
timestamp_end: None
quantify_dataset_version: 2.0.0
software_versions: {}
relationships: []
json_serialize_exclude: []And as before, we can reshape the dataset to take advantage of the xarray built-in utilities.
dataset_gridded = dh.to_gridded_dataset(
dataset_rep,
dimension="main_dim",
coords_names=dattrs.get_main_coords(dataset),
)
dataset_gridded
<xarray.Dataset> Size: 97kB
Dimensions: (amp: 30, time: 40, repetitions: 5)
Coordinates:
* amp (amp) float64 240B 0.45 0.4534 0.4569 ... 0.5431 0.5466 0.55
* time (time) float64 320B 0.0 2.564e-09 5.128e-09 ... 9.744e-08 1e-07
* repetitions (repetitions) <U1 20B 'A' 'B' 'C' 'D' 'E'
Data variables:
pop_q0 (repetitions, amp, time) float64 48kB 0.5 0.5 0.5 ... 0.5 0.5
pop_q1 (repetitions, amp, time) float64 48kB 0.5 0.5 0.5 ... 0.5 0.5
Attributes:
tuid: 20241014-175532-795-90ad16
dataset_name:
dataset_state: None
timestamp_start: None
timestamp_end: None
quantify_dataset_version: 2.0.0
software_versions: {}
relationships: []
json_serialize_exclude: []It is now possible to retrieve (select) a specific entry along the repetitions dimension:
_ = dataset_gridded.pop_q0.sel(repetitions="A").plot(x="amp")
plt.show()
_ = dataset_gridded.pop_q0.sel(repetitions="D").plot(x="amp")
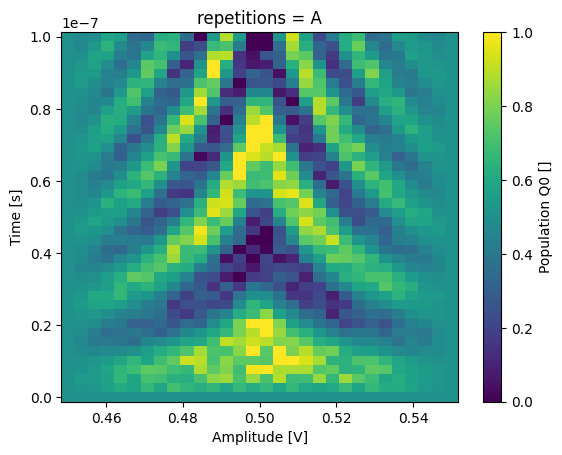
Dataset attributes#
The required attributes of the Quantify dataset are defined by the following dataclass.
It can be used to generate a default dictionary that is attached to a dataset under the xarray.Dataset.attrs attribute.
- class QDatasetAttrs(tuid=None, dataset_name='', dataset_state=None, timestamp_start=None, timestamp_end=None, quantify_dataset_version='2.0.0', software_versions=<factory>, relationships=<factory>, json_serialize_exclude=<factory>)[source]
Bases:
DataClassJsonMixinA dataclass representing the
attrsattribute of the Quantify dataset.See also
- dataset_name: str = ''
The dataset name, usually same as the the experiment name included in the name of the experiment container.
- dataset_state: Literal[None, 'running', 'interrupted (safety)', 'interrupted (forced)', 'done'] = None
Denotes the last known state of the experiment/data acquisition that served to ‘build’ this dataset. Can be used later to filter ‘bad’ datasets.
- json_serialize_exclude: List[str]
A list of strings corresponding to the names of other attributes that should not be json-serialized when writing the dataset to disk. Empty by default.
- quantify_dataset_version: str = '2.0.0'
A string identifying the version of this Quantify dataset for backwards compatibility.
- relationships: List[QDatasetIntraRelationship]
A list of relationships within the dataset specified as list of dictionaries that comply with the
QDatasetIntraRelationship.
- software_versions: Dict[str, str]
A mapping of other relevant software packages that are relevant to log for this dataset. Another example is the git tag or hash of a commit of a lab repository.
- timestamp_end: Union[str, None] = None
Human-readable timestamp (ISO8601) as returned by
datetime.datetime.now().astimezone().isoformat(). Specifies when the experiment/data acquisition ended.
- timestamp_start: Union[str, None] = None
Human-readable timestamp (ISO8601) as returned by
datetime.datetime.now().astimezone().isoformat(). Specifies when the experiment/data acquisition started.
- tuid: Union[str, None] = None
The time-based unique identifier of the dataset. See
quantify_core.data.types.TUID.
Additionally in order to express relationships between coordinates and/or variables the following template is provided:
- class QDatasetIntraRelationship(item_name=None, relation_type=None, related_names=<factory>, relation_metadata=<factory>)[source]
Bases:
DataClassJsonMixinA dataclass representing a dictionary that specifies a relationship between dataset variables.
A prominent example are calibration points contained within one variable or several variables that are necessary to interpret correctly the data of another variable.
- item_name: str | None = None
The name of the coordinate/variable to which we want to relate other coordinates/variables.
- related_names: List[str]
A list of names related to the
item_name.
- relation_metadata: Dict[str, Any]
A free-form dictionary to store additional information relevant to this relationship.
- relation_type: str | None = None
A string specifying the type of relationship.
Reserved relation types:
"calibration"- Specifies a list of main variables used as calibration data for the main variables whose name is specified by theitem_name.
from quantify_core.data.dataset_attrs import QDatasetAttrs
# tip: to_json and from_dict, from_json are also available
dataset.attrs = QDatasetAttrs().to_dict()
dataset.attrs
{
'tuid': None,
'dataset_name': '',
'dataset_state': None,
'timestamp_start': None,
'timestamp_end': None,
'quantify_dataset_version': '2.0.0',
'software_versions': {},
'relationships': [],
'json_serialize_exclude': []
}
Note that xarray automatically provides the entries of the dataset attributes as python attributes. And similarly for the xarray coordinates and data variables.
dataset.quantify_dataset_version, dataset.tuid
('2.0.0', None)
Main coordinates and variables attributes#
Similar to the dataset attributes (xarray.Dataset.attrs), the main coordinates and variables have each their own required attributes attached to them as a dictionary under the xarray.DataArray.attrs attribute.
- class QCoordAttrs(unit='', long_name='', is_main_coord=None, uniformly_spaced=None, is_dataset_ref=False, json_serialize_exclude=<factory>)[source]
Bases:
DataClassJsonMixinA dataclass representing the required
attrsattribute of main and secondary coordinates.- is_dataset_ref: bool = False
Flags if it is an array of
quantify_core.data.types.TUIDs of other dataset.
- is_main_coord: bool | None = None
When set to
True, flags the xarray coordinate to correspond to a main coordinate, otherwise (False) it corresponds to a secondary coordinate.
- json_serialize_exclude: List[str]
A list of strings corresponding to the names of other attributes that should not be json-serialized when writing the dataset to disk. Empty by default.
- long_name: str = ''
A long name for this coordinate.
- uniformly_spaced: bool | None = None
Indicates if the values are uniformly spaced.
- unit: str = ''
The units of the values.
dataset.amp.attrs
{
'unit': 'V',
'long_name': 'Amplitude',
'is_main_coord': True,
'uniformly_spaced': True,
'is_dataset_ref': False,
'json_serialize_exclude': []
}
- class QVarAttrs(unit='', long_name='', is_main_var=None, uniformly_spaced=None, grid=None, is_dataset_ref=False, has_repetitions=False, json_serialize_exclude=<factory>)[source]
Bases:
DataClassJsonMixinA dataclass representing the required
attrsattribute of main and secondary variables.- grid: bool | None = None
Indicates if the variables data are located on a grid, which does not need to be uniformly spaced along all dimensions. In other words, specifies if the corresponding main coordinates are the ‘unrolled’ points (also known as ‘unstacked’) corresponding to a grid.
If
Truethan it is possible to usequantify_core.data.handling.to_gridded_dataset()to convert the variables to a ‘stacked’ version.
- has_repetitions: bool = False
Indicates that the outermost dimension of this variable is a repetitions dimension. This attribute is intended to allow easy programmatic detection of such dimension. It can be used, for example, to average along this dimension before an automatic live plotting or analysis.
- is_dataset_ref: bool = False
Flags if it is an array of
quantify_core.data.types.TUIDs of other dataset. See also Dataset for a “nested MeasurementControl” experiment.
- is_main_var: bool | None = None
When set to
True, flags this xarray data variable to correspond to a main variable, otherwise (False) it corresponds to a secondary variable.
- json_serialize_exclude: List[str]
A list of strings corresponding to the names of other attributes that should not be json-serialized when writing the dataset to disk. Empty by default.
- long_name: str = ''
A long name for this coordinate.
- uniformly_spaced: bool | None = None
Indicates if the values are uniformly spaced. This does not apply to ‘true’ main variables but, because a MultiIndex is not supported yet by xarray when writing to disk, some coordinate variables have to be stored as main variables instead.
- unit: str = ''
The units of the values.
dataset.pop_q0.attrs
{
'unit': '',
'long_name': 'Population Q0',
'is_main_var': True,
'uniformly_spaced': True,
'grid': True,
'is_dataset_ref': False,
'has_repetitions': True,
'json_serialize_exclude': []
}
Storage format#
The Quantify dataset is written to disk and loaded back making use of xarray-supported facilities. Internally we write and load to/from disk using:
display_source_code(dh.write_dataset)
display_source_code(dh.load_dataset)
def write_dataset(path: Path | str, dataset: xr.Dataset) -> None:
"""Writes a :class:`~xarray.Dataset` to a file with the `h5netcdf` engine.
Before writing the
:meth:`~quantify_core.data.dataset_adapters.AdapterH5NetCDF.adapt`
is applied.
To accommodate for complex-type numbers and arrays ``invalid_netcdf=True`` is used.
Parameters
----------
path
Path to the file including filename and extension
dataset
The :class:`~xarray.Dataset` to be written to file.
""" # pylint: disable=line-too-long
_xarray_numpy_bool_patch(dataset) # See issue #161 in quantify-core
# Only quantify_dataset_version=>2.0.0 requires the adapter
if "quantify_dataset_version" in dataset.attrs:
dataset = da.AdapterH5NetCDF.adapt(dataset)
dataset.to_netcdf(path, engine="h5netcdf", invalid_netcdf=True)
def load_dataset(
tuid: TUID,
datadir: Path | str | None = None,
name: str = DATASET_NAME,
) -> xr.Dataset:
"""Loads a dataset specified by a tuid.
.. tip::
This method also works when specifying only the first part of a
:class:`~quantify_core.data.types.TUID`.
.. note::
This method uses :func:`~.load_dataset` to ensure the file is closed after
loading as datasets are intended to be immutable after performing the initial
experiment.
Parameters
----------
tuid
A :class:`~quantify_core.data.types.TUID` string. It is also possible to specify
only the first part of a tuid.
datadir
Path of the data directory. If ``None``, uses :meth:`~get_datadir` to determine
the data directory.
name
Name of the dataset.
Returns
-------
:
The dataset.
Raises
------
FileNotFoundError
No data found for specified date.
"""
return load_dataset_from_path(_locate_experiment_file(tuid, datadir, name))
Note that we use the h5netcdf engine which is more permissive than the default NetCDF engine to accommodate arrays of complex numbers.
Note
Furthermore, in order to support a variety of attribute types (e.g. the None type) and shapes (e.g. nested dictionaries) in a seamless dataset round trip, some additional tooling is required. See source codes below that implements the two-way conversion adapter used by the functions shown above.
display_source_code(dadapters.AdapterH5NetCDF)
class AdapterH5NetCDF(DatasetAdapterBase):
"""
Quantify dataset adapter for the ``h5netcdf`` engine.
It has the functionality of adapting the Quantify dataset to a format compatible
with the ``h5netcdf`` xarray backend engine that is used to write and load the
dataset to/from disk.
.. warning::
The ``h5netcdf`` engine has minor issues when performing a two-way trip of the
dataset. The ``type`` of some attributes are not preserved. E.g., list- and
tuple-like objects are loaded as numpy arrays of ``dtype=object``.
"""
@classmethod
def adapt(cls, dataset: xr.Dataset) -> xr.Dataset:
"""
Serializes to JSON the dataset and variables attributes.
To prevent the JSON serialization for specific items, their names should be
listed under the attribute named ``json_serialize_exclude`` (for each ``attrs``
dictionary).
Parameters
----------
dataset
Dataset that needs to be adapted.
Returns
-------
:
Dataset in which the attributes have been replaced with their JSON strings
version.
"""
return cls._transform(dataset, vals_converter=json.dumps)
@classmethod
def recover(cls, dataset: xr.Dataset) -> xr.Dataset:
"""
Reverts the action of ``.adapt()``.
To prevent the JSON de-serialization for specific items, their names should be
listed under the attribute named ``json_serialize_exclude``
(for each ``attrs`` dictionary).
Parameters
----------
dataset
Dataset from which to recover the original format.
Returns
-------
:
Dataset in which the attributes have been replaced with their python objects
version.
"""
return cls._transform(dataset, vals_converter=json.loads)
@staticmethod
def attrs_convert(
attrs: dict,
inplace: bool = False,
vals_converter: Callable[Any, Any] = json.dumps,
) -> dict:
"""
Converts to/from JSON string the values of the keys which are not listed in the
``json_serialize_exclude`` list.
Parameters
----------
attrs
The input dictionary.
inplace
If ``True`` the values are replaced in place, otherwise a deepcopy of
``attrs`` is performed first.
"""
json_serialize_exclude = attrs.get("json_serialize_exclude", [])
attrs = attrs if inplace else deepcopy(attrs)
for attr_name, attr_val in attrs.items():
if attr_name not in json_serialize_exclude:
attrs[attr_name] = vals_converter(attr_val)
return attrs
@classmethod
def _transform(
cls, dataset: xr.Dataset, vals_converter: Callable[Any, Any] = json.dumps
) -> xr.Dataset:
dataset = xr.Dataset(
dataset,
attrs=cls.attrs_convert(
dataset.attrs, inplace=False, vals_converter=vals_converter
),
)
for var_name in dataset.variables.keys():
# The new dataset generated above has already a deepcopy of the attributes.
_ = cls.attrs_convert(
dataset[var_name].attrs, inplace=True, vals_converter=vals_converter
)
return dataset