Tutorial 5. Plot monitor#
In this tutorial we dive into the capabilities of the plot monitor. We will create a fictional device and showcase how the plot monitor can be used. Enjoy!
Before instantiating any instruments or starting a measurement we change the
directory in which the experiments are saved using the
set_datadir()
[get_datadir()] functions.
⚠️ Warning!
We recommend always setting the directory at the start of the python kernel and stick to a single common data directory for all notebooks/experiments within your measurement setup/PC.
The cell below sets a default data directory (~/quantify-data on Linux/macOS or
$env:USERPROFILE\\quantify-data on Windows) for tutorial purposes. Change it to your
desired data directory. The utilities to find/search/extract data only work if
all the experiment containers are located within the same directory.
set_datadir(default_datadir())
Data will be saved in:
/root/quantify-data
QCoDeS drivers for our instruments#
class Device(Instrument):
"""A dummy instrument."""
def __init__(self, name: str):
super().__init__(name=name)
self.add_parameter(name="amp_0", unit="A", parameter_class=ManualParameter)
self.add_parameter(name="amp_1", unit="A", parameter_class=ManualParameter)
self.add_parameter(name="offset", unit="A", parameter_class=ManualParameter)
self.add_parameter(
name="adc", label="ADC input", unit="V", get_cmd=self._get_dac_value
)
def _get_dac_value(self):
s1 = np.exp(-3 * self.amp_0()) * np.sin(self.amp_0() * 2 * np.pi * 3)
s2 = np.cos(self.amp_1() * 2 * np.pi * 2)
return s1 + s2 + rng.uniform(0, 0.2) + self.offset()
Instantiate the instruments#
meas_ctrl = MeasurementControl("meas_ctrl")
plotmon = PlotMonitor_pyqt("PlotMonitor")
meas_ctrl.instr_plotmon(plotmon.name)
device = Device("Device")
Overview#
There are 3 parameters in the PlotMonitor_pyqt that control the datasets being displayed.
Two main parameters determine the datasets being displayed: tuids and tuids_extra.
plotmon.tuids()
plotmon.tuids_extra()
[]
[]
The interface is the same for both. The parameters accept a list of tuids or an empty list to reset.
### Example of loading datasets onto the plot
#plotmon.tuids(["20201124-184709-137-8a5112", "20201124-184716-237-918bee"])
#plotmon.tuids_extra(["20201124-184722-988-0463d4", "20201124-184729-618-85970f"])
The difference is that the MeasurementControl uses tuids and overrides them when running measurements.
Note
All the datasets must have matching data variables (settables and gettables).
The third relevant parameter is the tuids_max_num. It accepts an integer that determines the maximum number of datasets that will be stored in tuids when the MeasurementControl is running.
plotmon.tuids_max_num()
3
Note
This parameter has no effect when setting the tuids manually.
Usage examples#
# set initial values to emulate the instrument state
device.amp_0(0.0)
device.amp_1(0.0)
device.offset(0.0)
n_pnts = 50
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
Starting iterative measurement...
plotmon.main_QtPlot
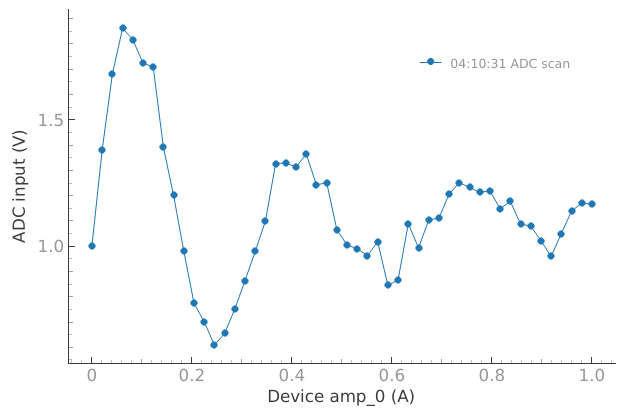
n_pnts = 20
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
Starting iterative measurement...
plotmon.main_QtPlot
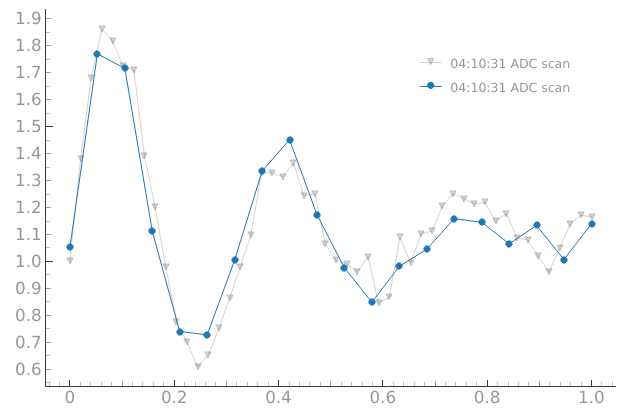
n_pnts = 30
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
Starting iterative measurement...
plotmon.main_QtPlot
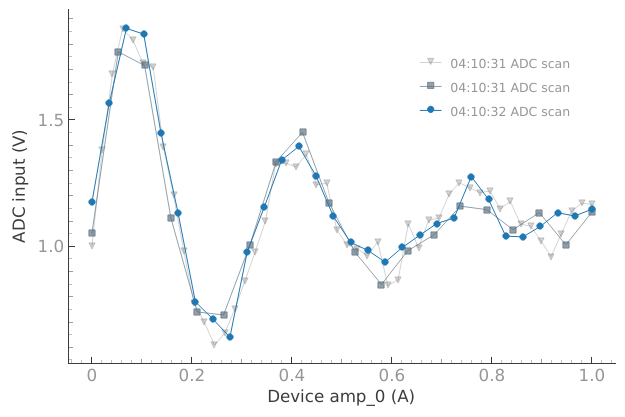
Now the oldest dataset will vanish from the plot:
# Now the oldest dataset will vanish from the plot
n_pnts = 40
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
Starting iterative measurement...
plotmon.main_QtPlot
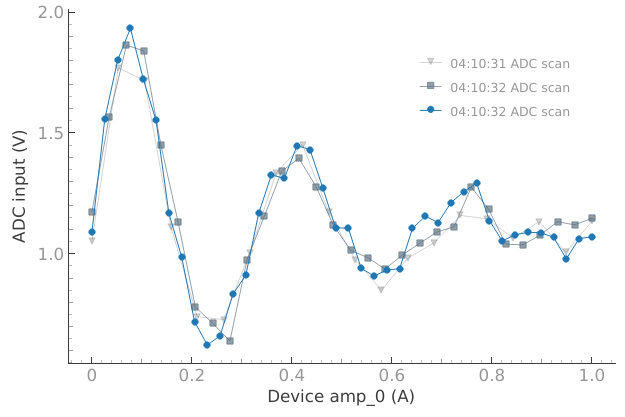
We can accumulate more datasets on the plot if we want to:
# We can accumulate more datasets on the plot if we want to
plotmon.tuids_max_num(4)
n_pnts = 40
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
Starting iterative measurement...
plotmon.main_QtPlot
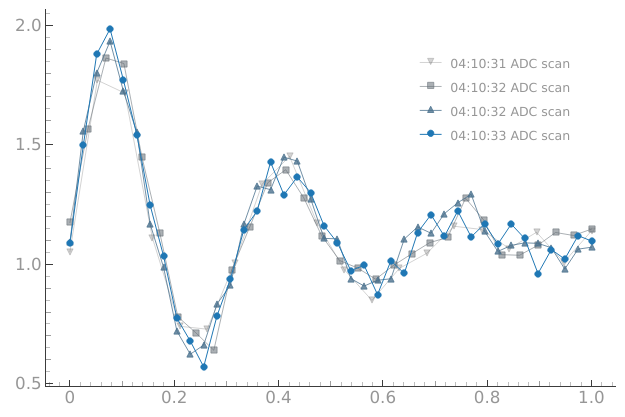
Or we can disable the accumulation and plot a single dataset:
# Or we can disable the accumulation and plot a single dataset
plotmon.tuids_max_num(1)
plotmon.main_QtPlot
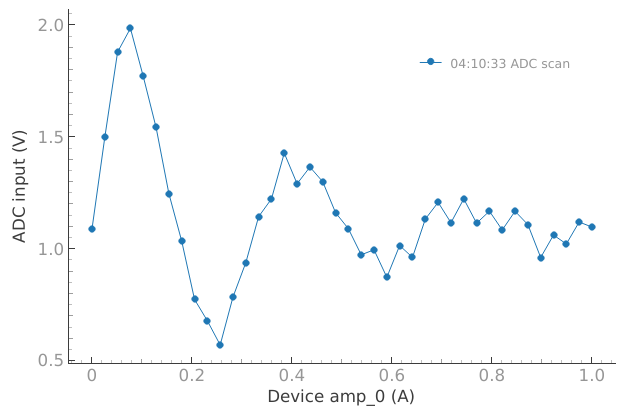
This can also be reset:
# This can also be reset with
plotmon.tuids([])
plotmon.main_QtPlot # The plotting window will vanish, it is supposed to

For now, we will allow two datasets on the plot monitor.
# For now we will allow two datasets on the plot monitor
plotmon.tuids_max_num(2)
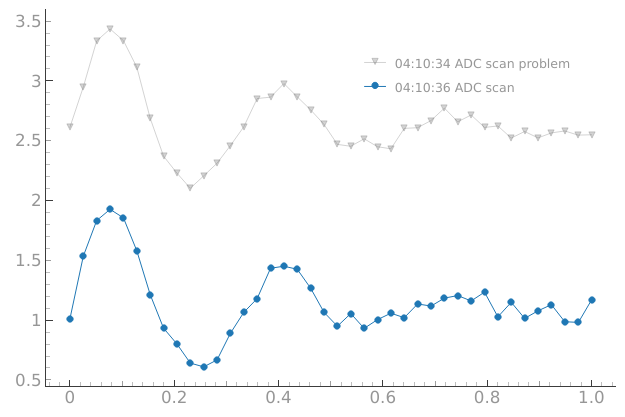
Now let’s imagine that something strange is happening with our setup…
# Now let's imagine that something strange is happening with our setup
device.offset(1.5)
n_pnts = 40
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan problem")
Starting iterative measurement...
plotmon.main_QtPlot
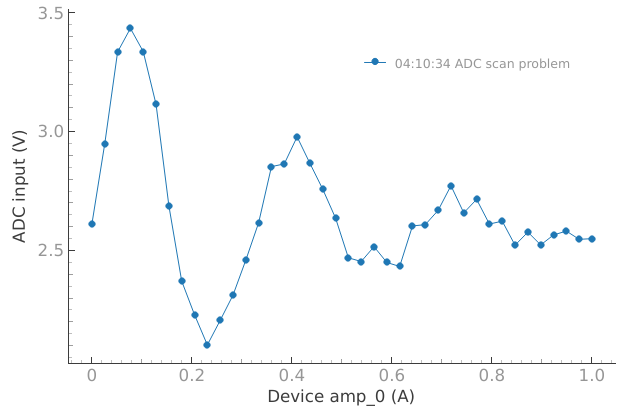
We would like to compare if the current behavior matches for example what we got a few minutes ago:
# We would like to compare if the current behavior matches for example
# what we got a few minutes ago
reference_tuids = sorted(get_tuids_containing("ADC"))[-3:-1]
plotmon.tuids_extra(reference_tuids)
plotmon.main_QtPlot
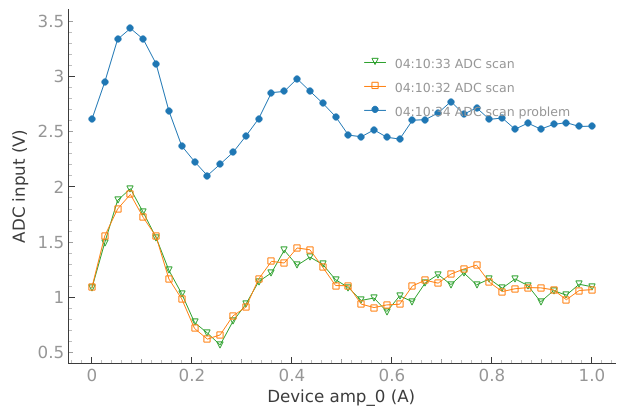
OK… that cable was not connected in the right place…
device.offset(0.0) # OK... that cable was not connected in the right place...
# Now let's run again our experiments while we compare it to the previous one in realtime
n_pnts = 30
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
n_pnts = 40
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
Starting iterative measurement...
Starting iterative measurement...
plotmon.main_QtPlot
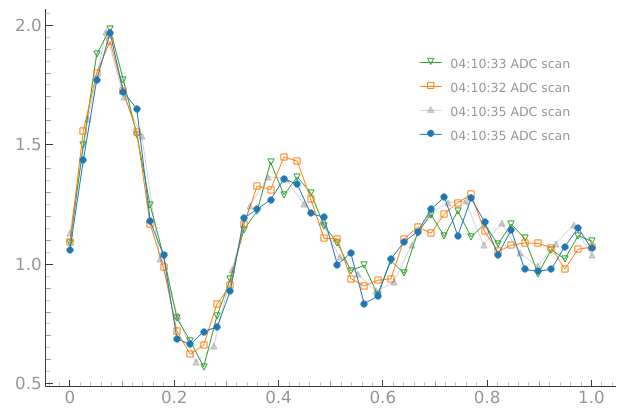
We do not need the reference datasets anymore
# We do not need the reference datasets anymore
plotmon.tuids_extra([])
plotmon.main_QtPlot
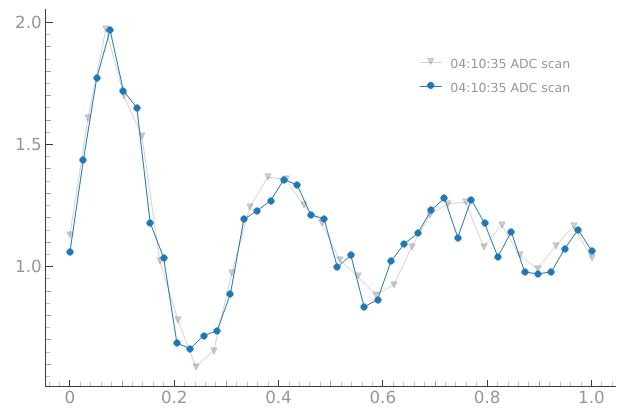
# Note: both plotmon.tuids_extra and plotmon.tuids can be used
# but keep in mind that meas_ctrl also uses the plotmon.tuids
tuids = get_tuids_containing("problem")[0:1]
tuids
plotmon.tuids(tuids)
n_pnts = 40
meas_ctrl.settables(device.amp_0)
meas_ctrl.setpoints(np.linspace(0, 1, n_pnts))
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan")
plotmon.main_QtPlot
['20250904-041034-293-08d17c']
Starting iterative measurement...
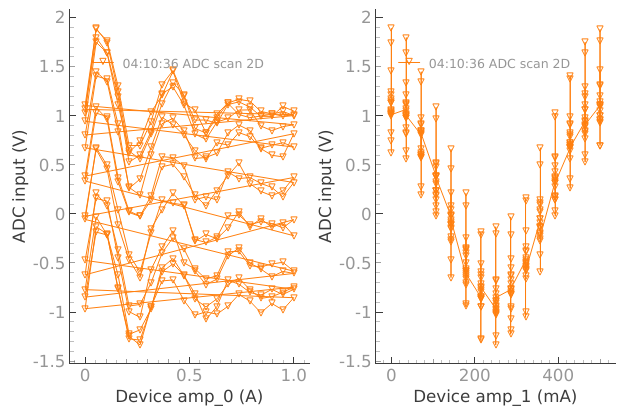
When we have 2D plots only the first dataset from plotmon.tuids or plotmon.tuids_extra will be plotted in the secondary window, in that order of priority.
# When we have 2D plots only the first dataset from plotmon.tuids or
# plotmon.tuids_extra, in that order of priority, will be plotted in the
# secondary window
meas_ctrl.settables([device.amp_0, device.amp_1])
meas_ctrl.setpoints_grid([np.linspace(0, 1, 20), np.linspace(0, 0.5, 15)])
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan 2D")
reference_tuid_2D = dset.attrs["tuid"]
Starting iterative measurement...
plotmon.main_QtPlot
plotmon.secondary_QtPlot
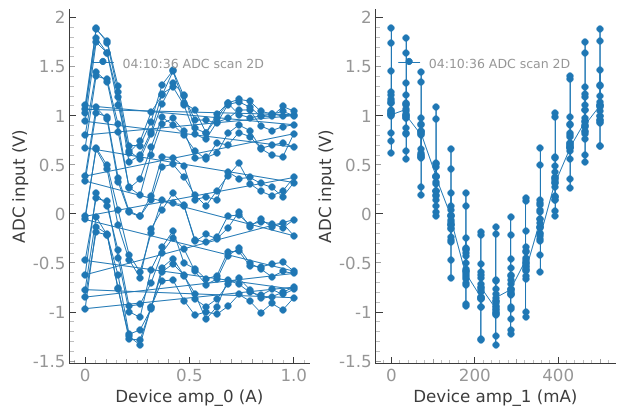
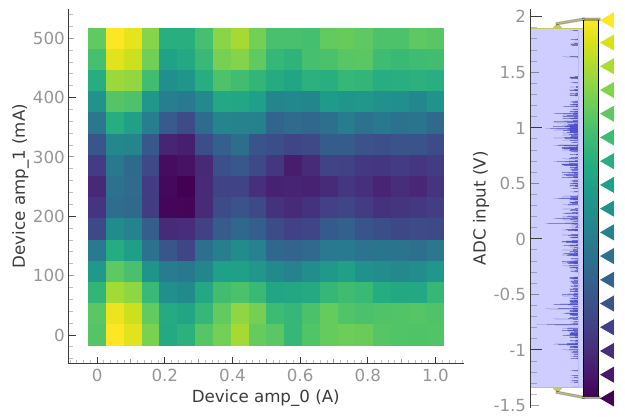
Note
The secondary window displays the last dataset with a 2D structure, and it remains persistent until replaced by a new dataset with a 2D structure.
Mind that the data on the secondary window does not always display data corresponding to the same dataset as the main window.
We still have the persistence of the previous dataset on the main window:
meas_ctrl.settables([device.amp_0, device.amp_1])
meas_ctrl.setpoints_grid([np.linspace(0, 1, 20), np.linspace(0, 0.5, 15)])
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan 2D")
Starting iterative measurement...
plotmon.main_QtPlot
plotmon.secondary_QtPlot
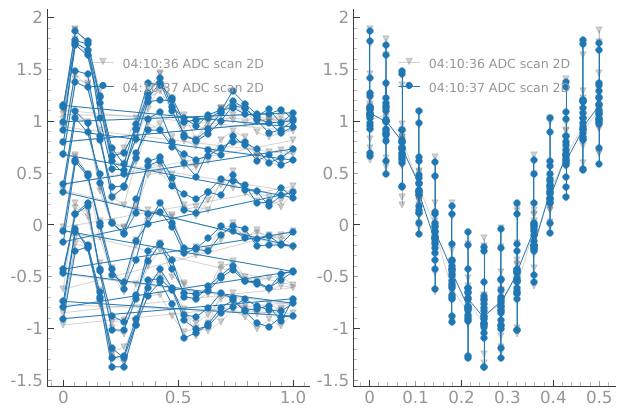
We can still have a permanent dataset as a reference in the main window:
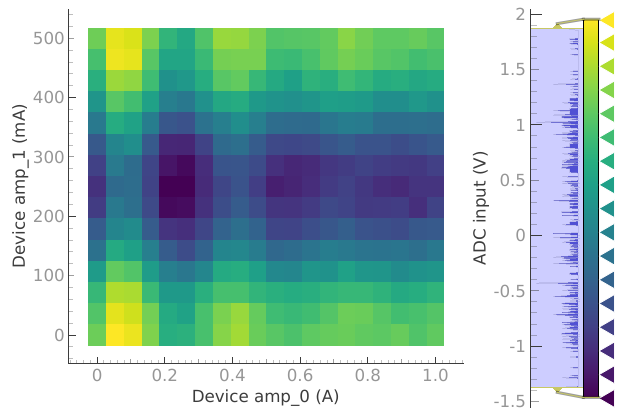
device.offset(2.03)
plotmon.tuids_extra([reference_tuid_2D])
meas_ctrl.settables([device.amp_0, device.amp_1])
meas_ctrl.setpoints_grid([np.linspace(0, 1, 20), np.linspace(0, 0.5, 15)])
meas_ctrl.gettables(device.adc)
dset = meas_ctrl.run("ADC scan 2D")
Starting iterative measurement...
plotmon.main_QtPlot
plotmon.secondary_QtPlot
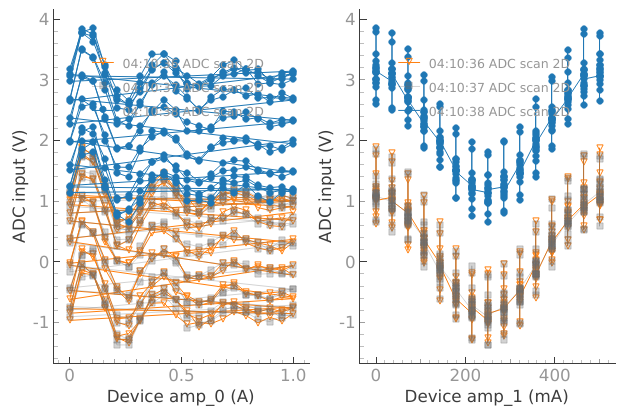
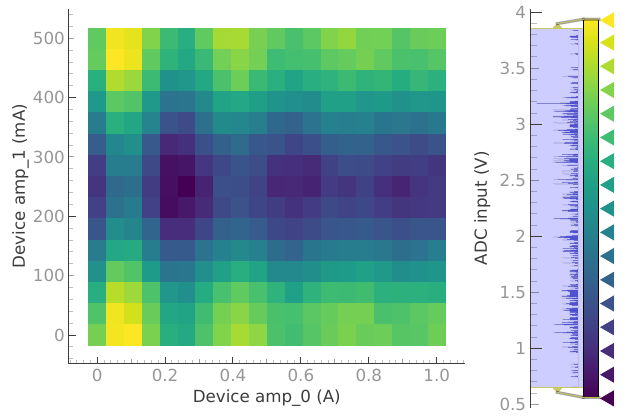
But if we want to see the 2D plot we need to reset plotmon.tuids.
plotmon.tuids([])
plotmon.tuids_extra([reference_tuid_2D])
plotmon.main_QtPlot
plotmon.secondary_QtPlot

plotmon.tuids()
[]
Now your life will never be the same again ;)