quantify_core.analysis#
base_analysis#
Module containing the analysis abstract base class and several basic analyses.
- class AnalysisMeta(name, bases, namespace, /, **kwargs)[source]#
Bases:
ABCMetaMetaclass, whose purpose is to avoid storing large amount of figure in memory.
By convention, analysis object stores figures in
self.figs_mplandself.axs_mpldictionaries. This causes troubles for long-running operations, because figures are all in memory and eventually this uses all available memory of the PC. In order to avoid it,BaseAnalysis.create_figures()and its derivatives are patched so that all the figures are put in LRU cache and reconstructed upon request toBaseAnalysis.figs_mplorBaseAnalysis.axs_mplif they were removed from the cache.Provided that analyses subclasses follow convention of figures being created in
BaseAnalysis.create_figures(), this approach should solve the memory issue and preserve reverse compatibility with present code.
- class AnalysisSteps(value)[source]#
Bases:
EnumAn enumerate of the steps executed by the
BaseAnalysis(and the default for subclasses).The involved steps are:
AnalysisSteps.STEP_1_PROCESS_DATA(BaseAnalysis.process_data())AnalysisSteps.STEP_2_RUN_FITTING(BaseAnalysis.run_fitting())AnalysisSteps.STEP_3_ANALYZE_FIT_RESULTS(BaseAnalysis.analyze_fit_results())AnalysisSteps.STEP_4_CREATE_FIGURES(BaseAnalysis.create_figures())AnalysisSteps.STEP_5_ADJUST_FIGURES(BaseAnalysis.adjust_figures())AnalysisSteps.STEP_6_SAVE_FIGURES(BaseAnalysis.save_figures())AnalysisSteps.STEP_7_SAVE_QUANTITIES_OF_INTEREST(BaseAnalysis.save_quantities_of_interest())AnalysisSteps.STEP_8_SAVE_PROCESSED_DATASET(BaseAnalysis.save_processed_dataset())AnalysisSteps.STEP_9_SAVE_FIT_RESULTS(BaseAnalysis.save_fit_results())
A custom analysis flow (e.g. inserting new steps) can be created by implementing an object similar to this one and overriding the
analysis_steps.
- class BaseAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
objectA template for analysis classes.
- analysis_steps#
Defines the steps of the analysis specified as an
Enum. Can be overridden in a subclass in order to define a custom analysis flow. SeeAnalysisStepsfor a template.alias of
AnalysisSteps
- __init__(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Initializes the variables used in the analysis and to which data is stored.
Warning
We highly discourage overriding the class initialization. If the analysis requires the user passing in any arguments, the
run()should be overridden and extended (see its docstring for an example).Settings schema:
Base analysis settings
properties
mpl_dpi
Matplotlib figures DPI.
type
integer
mpl_exclude_fig_titles
If
Truemaplotlib figures will not include the title.type
boolean
mpl_transparent_background
If
Truemaplotlib figures will have transparent background (when applicable).type
boolean
mpl_fig_formats
List of formats in which matplotlib figures will be saved. E.g.
['svg']type
array
items
type
string
save_fit_results
If True, save the results of fits in the Analysis sub-folder
type
boolean
- Parameters:
dataset (xr.Dataset | None (default:
None)) – an unprocessed (raw) quantify dataset to perform the analysis on.tuid (TUID | str | None (default:
None)) – if no dataset is specified, will look for the dataset with the matching tuid in the data directory.label (str (default:
'')) – if no dataset and no tuid is provided, will look for the most recent dataset that contains “label” in the name.settings_overwrite (dict | None (default:
None)) – A dictionary containing overrides for the global base_analysis.settings for this specific instance. See Settings schema above for available settings.plot_figures (bool (default:
True)) – Option to create and save figures for analysis.
- static _get_analysis_dir(tuid, name, create_missing=True)[source]#
Generate an analysis dir based on a given tuid and analysis class name.
- static _get_results_dir(analysis_dir, create_missing=True)[source]#
Generate an results dir based on a given analysis dir path.
- _repr_html_()[source]#
An html representation of the analysis class.
Shows the name of the analysis and TUID as well as the (.svg) figures generated by this analysis.
- adjust_clim(vmin, vmax, ax_ids=None)[source]#
Adjust the clim of matplotlib figures generated by analysis object.
- Parameters:
vmin (
float) – The bottom vlim in data coordinates. PassingNoneleaves the limit unchanged.vmax (
float) – The top vlim in data coordinates. Passing None leaves the limit unchanged.ax_ids (
Optional[list[str]] (default:None)) – A list of ax_ids specifying what axes to adjust. Passing None results in all axes of an analysis object being adjusted.
- Return type:
- adjust_cmap(cmap, ax_ids=None)[source]#
Adjust the cmap of matplotlib figures generated by analysis object.
- Parameters:
cmap (Colormap | str | None) – The colormap to set for the axis
ax_ids (list[str] (default:
None)) – A list of ax_ids specifying what axes to adjust. Passing None results in all axes of an analysis object being adjusted.
- adjust_figures()[source]#
Perform global adjustments after creating the figures but before saving them.
By default applies mpl_exclude_fig_titles and mpl_transparent_background from
.settings_overwriteto any matplotlib figures in.figs_mpl.Can be extended in a subclass for additional adjustments.
- adjust_xlim(xmin=None, xmax=None, ax_ids=None)[source]#
Adjust the xlim of matplotlib figures generated by analysis object.
- Parameters:
xmin (
Optional[float] (default:None)) – The bottom xlim in data coordinates. PassingNoneleaves the limit unchanged.xmax (
Optional[float] (default:None)) – The top xlim in data coordinates. Passing None leaves the limit unchanged.ax_ids (
Optional[list[str]] (default:None)) – A list of ax_ids specifying what axes to adjust. Passing None results in all axes of an analysis object being adjusted.
- Return type:
- adjust_ylim(ymin=None, ymax=None, ax_ids=None)[source]#
Adjust the ylim of matplotlib figures generated by analysis object.
- Parameters:
ymin (
Optional[float] (default:None)) – The bottom ylim in data coordinates. PassingNoneleaves the limit unchanged.ymax (
Optional[float] (default:None)) – The top ylim in data coordinates. Passing None leaves the limit unchanged.ax_ids (
Optional[list[str]] (default:None)) – A list of ax_ids specifying what axes to adjust. Passing None results in all axes of an analysis object being adjusted.
- Return type:
- analyze_fit_results()[source]#
To be implemented by subclasses.
Should analyze and process the
.fit_resultsand add the quantities of interest to the.quantities_of_interestdictionary.
- create_figures()[source]#
To be implemented by subclasses.
Should generate figures of interest. matplolib figures and axes objects should be added to the
.figs_mplandaxs_mpldictionaries., respectively.
- execute_analysis_steps()[source]#
Executes the methods corresponding to the analysis steps as defined by the
analysis_steps.Intended to be called by .run when creating a custom analysis that requires passing analysis configuration arguments to
run().
- extract_data()[source]#
If no dataset is provided, populates
.datasetwith data from the experiment matching the tuid/label.This method should be overwritten if an analysis does not relate to a single datafile.
- get_flow()[source]#
Returns a tuple with the ordered methods to be called by run analysis. Only return the figures methods if
self.plot_figuresisTrue.- Return type:
- classmethod load_fit_result(tuid, fit_name)[source]#
Load a saved
lmfit.model.ModelResultobject from file. For analyses that use custom fit functions, thecls.fit_function_definitionsobject must be defined in the subclass for that analysis.- Parameters:
- Return type:
- Returns:
: The lmfit model result object.
- process_data()[source]#
To be implemented by subclasses.
Should process, e.g., reshape, filter etc. the data before starting the analysis.
- run()[source]#
Execute analysis.
This function is at the core of all analysis. It calls
execute_analysis_steps()which executes all the methods defined in the.First step of any analysis is always extracting data, that is not configurable. Errors in extract_data() are considered fatal for analysis. Later steps are configurable by overriding
analysis_steps. Exceptions in these steps are logged and suppressed and analysis is considered partially successful.This function is typically called right after instantiating an analysis class.
Implementing a custom analysis that requires user input
When implementing your own custom analysis you might need to pass in a few configuration arguments. That should be achieved by overriding this function as show below.
from quantify_core.analysis.base_analysis import BaseAnalysis from typing_extensions import Self class MyAnalysis(BaseAnalysis): def run(self, optional_argument_one: float = 3.5e9) -> Self: # Save the value to be used in some step of the analysis self.optional_argument_one = optional_argument_one # Execute the analysis steps self.execute_analysis_steps() # Return the analysis object return self # ... other relevant methods ...
- Return type:
Self- Returns:
: The instance of the analysis object so that
run()returns an analysis object. You can initialize, run and assign it to a variable on a single line:, e.g.a_obj = MyAnalysis().run().
- run_fitting()[source]#
To be implemented by subclasses.
Should create fitting model(s) and fit data to the model(s) adding the result to the
.fit_resultsdictionary.
- save_figures()[source]#
Saves figures to disk. By default saves matplotlib figures.
Can be overridden or extended to make use of other plotting packages.
- save_figures_mpl(close_figs=True)[source]#
Saves all the matplotlib figures in the
.figs_mpldict.- Parameters:
close_figs (
bool(default:True)) – If True, closes matplotlib figures after saving.
- save_fit_results()[source]#
Saves the
lmfit.model.model_resultobjects for each fit in a sub-directory within the analysis directory.
- save_processed_dataset()[source]#
Saves a copy of the processed
.dataset_processedin the analysis folder of the experiment.
- save_quantities_of_interest()[source]#
Saves the
.quantities_of_interestas a JSON file in the analysis directory.The file is written using
json.dump()with theqcodes.utils.NumpyJSONEncodercustom encoder.
- property analysis_dir#
Analysis dir based on the tuid of the analysis class instance. Will create a directory if it does not exist.
- property name#
The name of the analysis, used in data saving.
- property results_dir#
Analysis dirrectory for this analysis. Will create a directory if it does not exist.
- class Basic1DAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BasicAnalysisDeprecated. Alias of
BasicAnalysisfor backwards compatibility.- run()[source]#
Execute analysis.
This function is at the core of all analysis. It calls
execute_analysis_steps()which executes all the methods defined in the.First step of any analysis is always extracting data, that is not configurable. Errors in extract_data() are considered fatal for analysis. Later steps are configurable by overriding
analysis_steps. Exceptions in these steps are logged and suppressed and analysis is considered partially successful.This function is typically called right after instantiating an analysis class.
Implementing a custom analysis that requires user input
When implementing your own custom analysis you might need to pass in a few configuration arguments. That should be achieved by overriding this function as show below.
from quantify_core.analysis.base_analysis import BaseAnalysis from typing_extensions import Self class MyAnalysis(BaseAnalysis): def run(self, optional_argument_one: float = 3.5e9) -> Self: # Save the value to be used in some step of the analysis self.optional_argument_one = optional_argument_one # Execute the analysis steps self.execute_analysis_steps() # Return the analysis object return self # ... other relevant methods ...
- Return type:
- Returns:
: The instance of the analysis object so that
run()returns an analysis object. You can initialize, run and assign it to a variable on a single line:, e.g.a_obj = MyAnalysis().run().
- class Basic2DAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisA basic analysis that extracts the data from the latest file matching the label and plots and stores the data in the experiment container.
- class BasicAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisA basic analysis that extracts the data from the latest file matching the label and plots and stores the data in the experiment container.
- analysis_steps_to_str(analysis_steps, class_name='BaseAnalysis')[source]#
A utility for generating the docstring for the analysis steps.
- Parameters:
analysis_steps (
Enum) – AnEnumsimilar toquantify_core.analysis.base_analysis.AnalysisSteps.class_name (
str(default:'BaseAnalysis')) – The class name that has the analysis_steps methods and for which the analysis_steps are intended.
- Return type:
- Returns:
: A formatted string version of the analysis_steps and corresponding methods.
- check_lmfit(fit_res)[source]#
Check that lmfit was able to successfully return a valid fit, and give a warning if not.
The function looks at lmfit’s success parameter, and also checks whether the fit was able to obtain valid error bars on the fitted parameters.
- Parameters:
fit_res (
ModelResult) – TheModelResultobject output by lmfit- Return type:
- Returns:
: A warning message if there is a problem with the fit.
- flatten_lmfit_modelresult(model)[source]#
Flatten an lmfit model result to a dictionary in order to be able to save it to disk.
Notes
We use this method as opposed to
save_modelresult()as the correspondingload_modelresult()cannot handle loading data with a custom fit function.
- lmfit_par_to_ufloat(param)[source]#
Safe conversion of an
lmfit.parameter.Parametertouncertainties.ufloat(value, std_dev).This function is intended to be used in custom analyses to avoid errors when an lmfit fails and the stderr is
None.
- wrap_text(text, width=35, replace_whitespace=True, **kwargs)[source]#
A text wrapping (braking over multiple lines) utility.
Intended to be used with
plot_textbox()in order to avoid too wide figure when, e.g.,check_lmfit()fails and a warning message is generated.For usage see, for example, source code of
create_figures().- Parameters:
text (str | None) – The text string to be wrapped over several lines.
width (int (default:
35)) – Maximum line width in characters.replace_whitespace (bool (default:
True)) – Passed totextwrap.wrap()and documented here.kwargs – Any other keyword arguments to be passed to
textwrap.wrap().
- Return type:
str | None
- Returns:
: The wrapped text (or
Noneif text isNone).
- settings = {'mpl_dpi': 450, 'mpl_fig_formats': ['png', 'svg'], 'mpl_exclude_fig_titles': False, 'mpl_transparent_background': True, 'save_fit_results': True}#
For convenience the analysis framework provides a set of global settings.
For available settings see
BaseAnalysis. These can be overwritten for each instance of an analysis.Examples
>>> from quantify_core.analysis import base_analysis as ba ... ba.settings["mpl_dpi"] = 300 # set resolution of matplotlib figures
cosine_analysis#
Module containing an education example of an analysis subclass.
See Tutorial 3. Building custom analyses - the data analysis framework that guides you through the process of building this analysis.
- class CosineAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisExemplary analysis subclass that fits a cosine to a dataset.
- process_data()[source]#
In some cases, you might need to process the data, e.g., reshape, filter etc., before starting the analysis. This is the method where it should be done.
See
process_data()for an implementation example.
- run_fitting()[source]#
Fits a
CosineModelto the data.
spectroscopy_analysis#
- class QubitFluxSpectroscopyAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAnalysis class for qubit flux spectroscopy.
Example
from quantify_core.analysis.spectroscopy_analysis import QubitFluxSpectroscopyAnalysis import quantify_core.data.handling as dh # load example data test_data_dir = "../tests/test_data" dh.set_datadir(test_data_dir) # run analysis and plot results analysis = ( QubitFluxSpectroscopyAnalysis(tuid="20230309-235354-353-9c94c5") .run() .display_figs_mpl() )
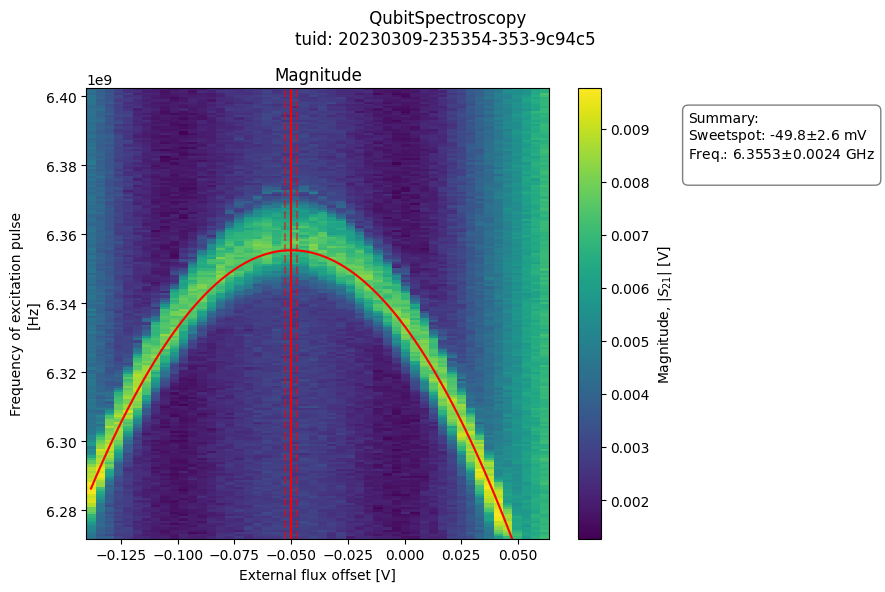
- analyze_fit_results()[source]#
Check the fit success and populate
.quantities_of_interest.- Return type:
- create_figures()[source]#
Generate plot of magnitude and phase images, with superposed model fit.
- Return type:
- class QubitSpectroscopyAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAnalysis for a qubit spectroscopy experiment.
Fits a Lorentzian function to qubit spectroscopy data and finds the 0-1 transistion frequency.
- class ResonatorFluxSpectroscopyAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAnalysis class for resonator flux spectroscopy.
Example
from quantify_core.analysis.spectroscopy_analysis import ( ResonatorFluxSpectroscopyAnalysis ) import quantify_core.data.handling as dh # load example data test_data_dir = "../tests/test_data" dh.set_datadir(test_data_dir) # run analysis and plot results analysis = ( ResonatorFluxSpectroscopyAnalysis(tuid="20230308-235659-059-cf471e") .run() .display_figs_mpl() )
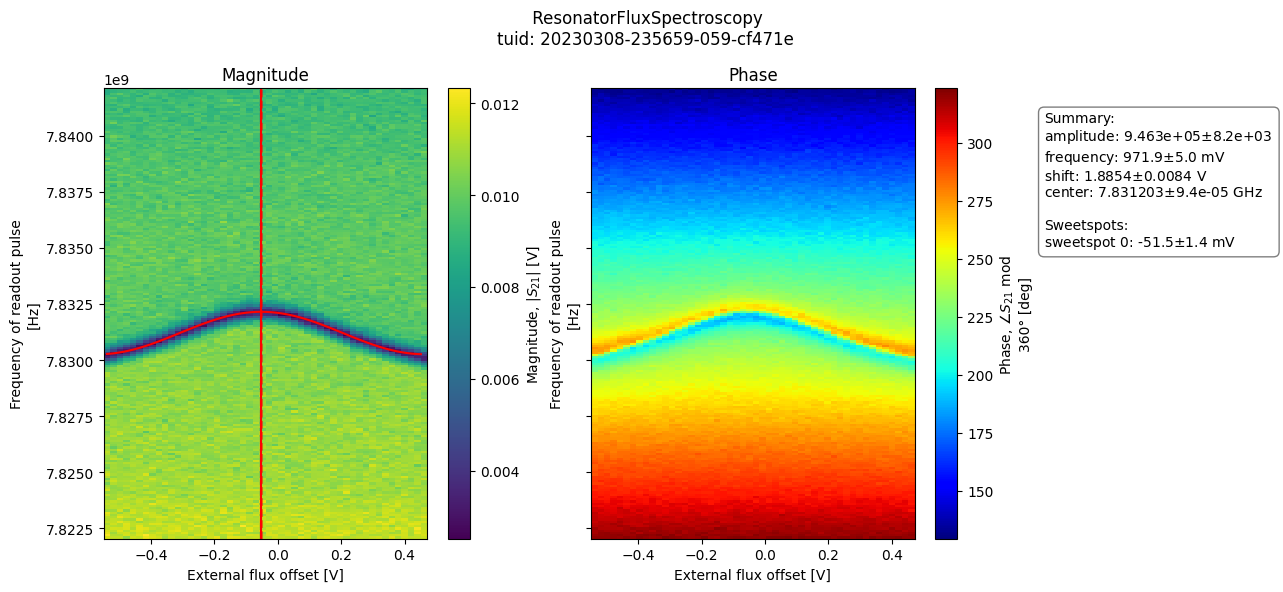
- analyze_fit_results()[source]#
Check the fit success and populate
.quantities_of_interest.- Return type:
- create_figures()[source]#
Generate plot of magnitude and phase images, with superposed model fit.
- Return type:
- class ResonatorSpectroscopyAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAnalysis for a spectroscopy experiment of a hanger resonator.
- create_figures()[source]#
Plots the measured and fitted transmission \(S_{21}\) as the I and Q component vs frequency, the magnitude and phase vs frequency, and on the complex I,Q plane.
- process_data()[source]#
Verifies that the data is measured as magnitude and phase and casts it to a dataset of complex valued transmission \(S_{21}\).
- run_fitting()[source]#
Fits a
ResonatorModelto the data.
single_qubit_timedomain#
Module containing analyses for common single qubit timedomain experiments.
- class AllXYAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
SingleQubitTimedomainAnalysisNormalizes the data from an AllXY experiment and plots against an ideal curve.
See section 2.3.2 of Reed [2013] for an explanation of the AllXY experiment and it’s applications in diagnosing errors in single-qubit control pulses.
- create_figures()[source]#
To be implemented by subclasses.
Should generate figures of interest. matplolib figures and axes objects should be added to the
.figs_mplandaxs_mpldictionaries., respectively.
- process_data()[source]#
Processes the data so that the analysis can make assumptions on the format.
Populates self.dataset_processed.S21 with the complex (I,Q) valued transmission, and if calibration points are present for the 0 and 1 state, populates self.dataset_processed.pop_exc with the excited state population.
- run()[source]#
Executes the analysis using specific datapoints as calibration points.
- Returns:
AllXYAnalysis: The instance of this analysis.
- class EchoAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
SingleQubitTimedomainAnalysis,_DecayFigMixinAnalysis class for a qubit spin-echo experiment, which fits an exponential decay and extracts the T2_echo time.
- run_fitting()[source]#
Fit the data to
ExpDecayModel.
- class RabiAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
SingleQubitTimedomainAnalysisFits a cosine curve to Rabi oscillation data and finds the qubit drive amplitude required to implement a pi-pulse.
The analysis will automatically rotate the data so that the data lies along the axis with the best SNR.
- _rotate_to_calibrated_axis()[source]#
If calibration points are True, automatically determine the point farthest from the 0 point to use as a reference to rotate the data.
This will ensure the data lies along the axis with the best SNR.
- run(calibration_points=True)[source]#
- Parameters:
calibration_points (
bool(default:True)) – Specifies if the data should be rotated so that it lies along the axis with the best SNR.- Returns:
RabiAnalysis: The instance of this analysis.
- class RamseyAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
SingleQubitTimedomainAnalysis,_DecayFigMixinFits a decaying cosine curve to Ramsey data (possibly with artificial detuning) and finds the true detuning, qubit frequency and T2* time.
- analyze_fit_results()[source]#
Extract the real detuning and qubit frequency based on the artificial detuning and fitted detuning.
- run(artificial_detuning=0, qubit_frequency=None, calibration_points='auto')[source]#
- Parameters:
artificial_detuning (
float(default:0)) – The detuning in Hz that will be emulated by adding an extra phase in software.qubit_frequency (
Optional[float] (default:None)) – The initial recorded value of the qubit frequency (before accurate fitting is done) in Hz.calibration_points (
Union[bool,Literal['auto']] (default:'auto')) – Indicates if the data analyzed includes calibration points. If set toTrue, will interpret the last two data points in the dataset as \(|0\rangle\) and \(|1\rangle\) respectively. If"auto", will usehas_calibration_points()to determine if the data contains calibration points.
- Returns:
RamseyAnalysis: The instance of this analysis.
- run_fitting()[source]#
Fits a
DecayOscillationModelto the data.
- class SingleQubitTimedomainAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisBase Analysis class for single-qubit timedomain experiments.
- process_data()[source]#
Processes the data so that the analysis can make assumptions on the format.
Populates self.dataset_processed.S21 with the complex (I,Q) valued transmission, and if calibration points are present for the 0 and 1 state, populates self.dataset_processed.pop_exc with the excited state population.
- run(calibration_points='auto')[source]#
- Parameters:
calibration_points (
Union[bool,Literal['auto']] (default:'auto')) – Indicates if the data analyzed includes calibration points. If set toTrue, will interpret the last two data points in the dataset as \(|0\rangle\) and \(|1\rangle\) respectively. If"auto", will usehas_calibration_points()to determine if the data contains calibration points.- Returns:
SingleQubitTimedomainAnalysis: The instance of this analysis.
- class T1Analysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
SingleQubitTimedomainAnalysis,_DecayFigMixinAnalysis class for a qubit T1 experiment, which fits an exponential decay and extracts the T1 time.
- run_fitting()[source]#
Fit the data to
ExpDecayModel.
interpolation_analysis#
- class InterpolationAnalysis2D(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAn analysis class which generates a 2D interpolating plot for each yi variable in the dataset.
optimization_analysis#
- class OptimizationAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAn analysis class which extracts the optimal quantities from an N-dimensional interpolating experiment.
- process_data()[source]#
Finds the optimal (minimum or maximum) for y0 and saves the xi and y0 values in the
quantities_of_interest.
- run(minimize=True)[source]#
- Parameters:
minimize (
bool(default:True)) – Boolean which determines whether to report the minimum or the maximum. True for minimize. False for maximize.- Returns:
OptimizationAnalysis: The instance of this analysis.
fitting_models#
Models and fit functions to be used with the lmfit fitting framework.
- class CosineModel(*args, **kwargs)[source]#
Bases:
ModelExemplary lmfit model with a guess for a cosine.
Note
The
lmfit.modelsmodule provides several fitting models that might fit your needs out of the box.- __init__(*args, **kwargs)[source]#
- Parameters:
independent_vars (
listofstr) – Arguments to the model function that are independent variables default is['x']).prefix (
str) – String to prepend to parameter names, needed to add two Models that have parameter names in common.nan_policy – How to handle NaN and missing values in data. See Notes below.
**kwargs – Keyword arguments to pass to
Model.
Notes
1. nan_policy sets what to do when a NaN or missing value is seen in the data. Should be one of:
‘raise’ : raise a ValueError (default)
‘propagate’ : do nothing
‘omit’ : drop missing data
See also
- guess(data, x, **kws)[source]#
Guess starting values for the parameters of a model.
- Parameters:
- Return type:
- Returns:
- params
Parameters Initial, guessed values for the parameters of a Model.
Changed in version 1.0.3: Argument
xis now explicitly required to estimate starting values.- params
- class DecayOscillationModel(*args, **kwargs)[source]#
Bases:
ModelModel for a decaying oscillation which decays to a point with 0 offset from the centre of the of the oscillation (as in a Ramsey experiment, for example).
- __init__(*args, **kwargs)[source]#
- Parameters:
independent_vars (
listofstr) – Arguments to the model function that are independent variables default is['x']).prefix (
str) – String to prepend to parameter names, needed to add two Models that have parameter names in common.nan_policy – How to handle NaN and missing values in data. See Notes below.
**kwargs – Keyword arguments to pass to
Model.
Notes
1. nan_policy sets what to do when a NaN or missing value is seen in the data. Should be one of:
‘raise’ : raise a ValueError (default)
‘propagate’ : do nothing
‘omit’ : drop missing data
See also
- guess(data, **kws)[source]#
Guess starting values for the parameters of a model.
- Parameters:
- Return type:
- Returns:
- params
Parameters Initial, guessed values for the parameters of a Model.
Changed in version 1.0.3: Argument
xis now explicitly required to estimate starting values.- params
- class ExpDecayModel(*args, **kwargs)[source]#
Bases:
ModelModel for an exponential decay, such as a qubit T1 measurement.
- __init__(*args, **kwargs)[source]#
- Parameters:
independent_vars (
listofstr) – Arguments to the model function that are independent variables default is['x']).prefix (
str) – String to prepend to parameter names, needed to add two Models that have parameter names in common.nan_policy – How to handle NaN and missing values in data. See Notes below.
**kwargs – Keyword arguments to pass to
Model.
Notes
1. nan_policy sets what to do when a NaN or missing value is seen in the data. Should be one of:
‘raise’ : raise a ValueError (default)
‘propagate’ : do nothing
‘omit’ : drop missing data
See also
- guess(data, **kws)[source]#
Guess starting values for the parameters of a model.
- Parameters:
- Return type:
- Returns:
- params
Parameters Initial, guessed values for the parameters of a Model.
Changed in version 1.0.3: Argument
xis now explicitly required to estimate starting values.- params
- class LorentzianModel(*args, **kwargs)[source]#
Bases:
ModelModel for data which follows a Lorentzian function.
Uses the function
lorentzian_func()as the defining equation.
- class RabiModel(*args, **kwargs)[source]#
Bases:
ModelModel for a Rabi oscillation as a function of the microwave drive amplitude. Phase of oscillation is fixed at \(\pi\) in order to ensure that the oscillation is at a minimum when the drive amplitude is 0.
- __init__(*args, **kwargs)[source]#
- Parameters:
independent_vars (
listofstr) – Arguments to the model function that are independent variables default is['x']).prefix (
str) – String to prepend to parameter names, needed to add two Models that have parameter names in common.nan_policy – How to handle NaN and missing values in data. See Notes below.
**kwargs – Keyword arguments to pass to
Model.
Notes
1. nan_policy sets what to do when a NaN or missing value is seen in the data. Should be one of:
‘raise’ : raise a ValueError (default)
‘propagate’ : do nothing
‘omit’ : drop missing data
See also
- guess(data, **kws)[source]#
Guess starting values for the parameters of a model.
- Parameters:
- Return type:
- Returns:
- params
Parameters Initial, guessed values for the parameters of a Model.
Changed in version 1.0.3: Argument
xis now explicitly required to estimate starting values.- params
- class ResonatorModel(*args, **kwargs)[source]#
Bases:
ModelResonator model
Implementation and design patterns inspired by the complex resonator model example (lmfit documentation).
- __init__(*args, **kwargs)[source]#
- Parameters:
independent_vars (
listofstr) – Arguments to the model function that are independent variables default is['x']).prefix (
str) – String to prepend to parameter names, needed to add two Models that have parameter names in common.nan_policy – How to handle NaN and missing values in data. See Notes below.
**kwargs – Keyword arguments to pass to
Model.
Notes
1. nan_policy sets what to do when a NaN or missing value is seen in the data. Should be one of:
‘raise’ : raise a ValueError (default)
‘propagate’ : do nothing
‘omit’ : drop missing data
See also
- guess(data, **kws)[source]#
Guess starting values for the parameters of a model.
- Parameters:
- Return type:
- Returns:
- params
Parameters Initial, guessed values for the parameters of a Model.
Changed in version 1.0.3: Argument
xis now explicitly required to estimate starting values.- params
- cos_func(x, frequency, amplitude, offset, phase=0)[source]#
An oscillating cosine function:
\(y = \mathrm{amplitude} \times \cos(2 \pi \times \mathrm{frequency} \times x + \mathrm{phase}) + \mathrm{offset}\)
- Parameters:
- Return type:
- Returns:
: Output signal magnitude
- exp_damp_osc_func(t, tau, n_factor, frequency, phase, amplitude, offset)[source]#
A sinusoidal oscillation with an exponentially decaying envelope function:
\(y = \mathrm{amplitude} \times \exp\left(-(t/\tau)^\mathrm{n\_factor}\right)(\cos(2\pi\mathrm{frequency}\times t + \mathrm{phase}) + \mathrm{oscillation_offset}) + \mathrm{exponential_offset}\)
- Parameters:
t (
float) – timetau (
float) – decay timen_factor (
float) – exponential decay factorfrequency (
float) – frequency of the oscillationphase (
float) – phase of the oscillationamplitude (
float) – initial amplitude of the oscillationoscillation_offset – vertical offset of cosine oscillation relative to exponential asymptote
exponential_offset – offset of exponential asymptote
- Returns:
: Output of decaying cosine function as a float
- exp_decay_func(t, tau, amplitude, offset, n_factor)[source]#
This is a general exponential decay function:
\(y = \mathrm{amplitude} \times \exp\left(-(t/\tau)^\mathrm{n\_factor}\right) + \mathrm{offset}\)
- Parameters:
- Return type:
- Returns:
: Output of exponential function as a float
- fft_freq_phase_guess(data, t)[source]#
Guess for a cosine fit using FFT, only works for evenly spaced points.
- get_guess_common_doc()[source]#
Returns a common docstring to be used for the
guess()method of custom fittingModels. :rtype:strUsage example for a custom fitting model
See the usage example at the end of the
ResonatorModelsource-code:class ResonatorModel(lmfit.model.Model): """ Resonator model Implementation and design patterns inspired by the `complex resonator model example <https://lmfit.github.io/lmfit-py/examples/example_complex_resonator_model.html>`_ (`lmfit` documentation). """ # pylint: disable=line-too-long # pylint: disable=empty-docstring # pylint: disable=abstract-method def __init__(self, *args, **kwargs): # pass in the defining equation so the user doesn't have to later. super().__init__(hanger_func_complex_SI, *args, **kwargs) self.set_param_hint("Ql", min=0) # Enforce Q is positive self.set_param_hint("Qe", min=0) # Enforce Q is positive # Internal and coupled quality factor can be derived from fitted params self.set_param_hint("Qi", expr="1./(1./Ql-1./Qe*cos(theta))", vary=False) self.set_param_hint("Qc", expr="Qe/cos(theta)", vary=False) # pylint: disable=too-many-locals # pylint: disable=missing-function-docstring def guess(self, data, **kws) -> lmfit.parameter.Parameters: f = kws.get("f") if f is None: return None argmin_s21 = np.abs(data).argmin() fmin = f.min() fmax = f.max() # guess that the resonance is the lowest point fr_guess = f[argmin_s21] # assume the user isn't trying to fit just a small part of a resonance curve. Q_min = 0.1 * (fr_guess / (fmax - fmin)) delta_f = np.diff(f) # assume f is sorted min_delta_f = delta_f[delta_f > 0].min() Q_max = ( fr_guess / min_delta_f ) # assume data actually samples the resonance reasonably Q_guess = np.sqrt(Q_min * Q_max) # geometric mean, why not? (phi_0_guess, phi_v_guess) = resonator_phase_guess( data, f ) # Come up with a guess for phase velocity self.set_param_hint("fr", value=fr_guess, min=fmin, max=fmax) self.set_param_hint("Ql", value=Q_guess * 1.01, min=Q_min, max=Q_max) self.set_param_hint("Qe", value=Q_guess * 0.99, min=0) self.set_param_hint("A", value=np.mean(abs(data)), min=0) # The parameters below need a proper guess. self.set_param_hint("theta", value=0, min=-np.pi / 2, max=np.pi / 2) self.set_param_hint("phi_0", value=phi_0_guess) self.set_param_hint("phi_v", value=phi_v_guess) self.set_param_hint("alpha", value=0, min=-1, max=1) params = self.make_params() return lmfit.models.update_param_vals(params, self.prefix, **kws) # Same design patter is used in lmfit.models __init__.__doc__ = get_model_common_doc() + mk_seealso("hanger_func_complex_SI") guess.__doc__ = get_guess_common_doc()
- get_model_common_doc()[source]#
Returns a common docstring to be used with custom fitting
Models. :rtype:strUsage example for a custom fitting model
See the usage example at the end of the
ResonatorModelsource-code:class ResonatorModel(lmfit.model.Model): """ Resonator model Implementation and design patterns inspired by the `complex resonator model example <https://lmfit.github.io/lmfit-py/examples/example_complex_resonator_model.html>`_ (`lmfit` documentation). """ # pylint: disable=line-too-long # pylint: disable=empty-docstring # pylint: disable=abstract-method def __init__(self, *args, **kwargs): # pass in the defining equation so the user doesn't have to later. super().__init__(hanger_func_complex_SI, *args, **kwargs) self.set_param_hint("Ql", min=0) # Enforce Q is positive self.set_param_hint("Qe", min=0) # Enforce Q is positive # Internal and coupled quality factor can be derived from fitted params self.set_param_hint("Qi", expr="1./(1./Ql-1./Qe*cos(theta))", vary=False) self.set_param_hint("Qc", expr="Qe/cos(theta)", vary=False) # pylint: disable=too-many-locals # pylint: disable=missing-function-docstring def guess(self, data, **kws) -> lmfit.parameter.Parameters: f = kws.get("f") if f is None: return None argmin_s21 = np.abs(data).argmin() fmin = f.min() fmax = f.max() # guess that the resonance is the lowest point fr_guess = f[argmin_s21] # assume the user isn't trying to fit just a small part of a resonance curve. Q_min = 0.1 * (fr_guess / (fmax - fmin)) delta_f = np.diff(f) # assume f is sorted min_delta_f = delta_f[delta_f > 0].min() Q_max = ( fr_guess / min_delta_f ) # assume data actually samples the resonance reasonably Q_guess = np.sqrt(Q_min * Q_max) # geometric mean, why not? (phi_0_guess, phi_v_guess) = resonator_phase_guess( data, f ) # Come up with a guess for phase velocity self.set_param_hint("fr", value=fr_guess, min=fmin, max=fmax) self.set_param_hint("Ql", value=Q_guess * 1.01, min=Q_min, max=Q_max) self.set_param_hint("Qe", value=Q_guess * 0.99, min=0) self.set_param_hint("A", value=np.mean(abs(data)), min=0) # The parameters below need a proper guess. self.set_param_hint("theta", value=0, min=-np.pi / 2, max=np.pi / 2) self.set_param_hint("phi_0", value=phi_0_guess) self.set_param_hint("phi_v", value=phi_v_guess) self.set_param_hint("alpha", value=0, min=-1, max=1) params = self.make_params() return lmfit.models.update_param_vals(params, self.prefix, **kws) # Same design patter is used in lmfit.models __init__.__doc__ = get_model_common_doc() + mk_seealso("hanger_func_complex_SI") guess.__doc__ = get_guess_common_doc()
- hanger_func_complex_SI(f, fr, Ql, Qe, A, theta, phi_v, phi_0, alpha=1)[source]#
This is the complex function for a hanger (lambda/4 resonator).
- Parameters:
f (
float) – frequencyfr (
float) – resonance frequencyA (
float) – background transmission amplitudeQl (
float) – loaded quality factor of the resonatorQe (
float) – magnitude of extrinsic quality factorQe = |Q_extrinsic|theta (
float) – phase of extrinsic quality factor (in rad)phi_v (
float) – phase to account for propagation delay to samplephi_0 (
float) – phase to account for propagation delay from samplealpha (
float(default:1)) – slope of signal around the resonance
- Return type:
- Returns:
: complex valued transmission
See eq. S4 from Bruno et al. (2015) ArXiv:1502.04082.
\[S_{21} = A \left(1+\alpha \frac{f-f_r}{f_r} \right) \left(1- \frac{\frac{Q_l}{|Q_e|}e^{i\theta} }{1+2iQ_l \frac{f-f_r}{f_r}} \right) e^{i (\phi_v f + \phi_0)}\]The loaded and extrinsic quality factors are related to the internal and coupled Q according to:
\[\frac{1}{Q_l} = \frac{1}{Q_c}+\frac{1}{Q_i}\]and
\[\frac{1}{Q_c} = \mathrm{Re}\left(\frac{1}{|Q_e|e^{-i\theta}}\right)\]
- lorentzian_func(x, x0, width, a, c)[source]#
A Lorentzian function.
\[y = \frac{a*\mathrm{width}}{\pi(\mathrm{width}^2 + (x - x_0)^2)} + c\]
- mk_seealso(function_name, role='func', prefix='\\n\\n', module_location='.')[source]#
Returns a sphinx seealso pointing to a function.
Intended to be used for building custom fitting model docstrings.
Usage example for a custom fitting model
See the usage example at the end of the
ResonatorModelsource-code:class ResonatorModel(lmfit.model.Model): """ Resonator model Implementation and design patterns inspired by the `complex resonator model example <https://lmfit.github.io/lmfit-py/examples/example_complex_resonator_model.html>`_ (`lmfit` documentation). """ # pylint: disable=line-too-long # pylint: disable=empty-docstring # pylint: disable=abstract-method def __init__(self, *args, **kwargs): # pass in the defining equation so the user doesn't have to later. super().__init__(hanger_func_complex_SI, *args, **kwargs) self.set_param_hint("Ql", min=0) # Enforce Q is positive self.set_param_hint("Qe", min=0) # Enforce Q is positive # Internal and coupled quality factor can be derived from fitted params self.set_param_hint("Qi", expr="1./(1./Ql-1./Qe*cos(theta))", vary=False) self.set_param_hint("Qc", expr="Qe/cos(theta)", vary=False) # pylint: disable=too-many-locals # pylint: disable=missing-function-docstring def guess(self, data, **kws) -> lmfit.parameter.Parameters: f = kws.get("f") if f is None: return None argmin_s21 = np.abs(data).argmin() fmin = f.min() fmax = f.max() # guess that the resonance is the lowest point fr_guess = f[argmin_s21] # assume the user isn't trying to fit just a small part of a resonance curve. Q_min = 0.1 * (fr_guess / (fmax - fmin)) delta_f = np.diff(f) # assume f is sorted min_delta_f = delta_f[delta_f > 0].min() Q_max = ( fr_guess / min_delta_f ) # assume data actually samples the resonance reasonably Q_guess = np.sqrt(Q_min * Q_max) # geometric mean, why not? (phi_0_guess, phi_v_guess) = resonator_phase_guess( data, f ) # Come up with a guess for phase velocity self.set_param_hint("fr", value=fr_guess, min=fmin, max=fmax) self.set_param_hint("Ql", value=Q_guess * 1.01, min=Q_min, max=Q_max) self.set_param_hint("Qe", value=Q_guess * 0.99, min=0) self.set_param_hint("A", value=np.mean(abs(data)), min=0) # The parameters below need a proper guess. self.set_param_hint("theta", value=0, min=-np.pi / 2, max=np.pi / 2) self.set_param_hint("phi_0", value=phi_0_guess) self.set_param_hint("phi_v", value=phi_v_guess) self.set_param_hint("alpha", value=0, min=-1, max=1) params = self.make_params() return lmfit.models.update_param_vals(params, self.prefix, **kws) # Same design patter is used in lmfit.models __init__.__doc__ = get_model_common_doc() + mk_seealso("hanger_func_complex_SI") guess.__doc__ = get_guess_common_doc()
- Parameters:
function_name (
str) – name of the function to point torole (
str(default:'func')) – a sphinx role, e.g."func"prefix (
str(default:'\\n\\n')) – string preceding the seealsomodule_location (
str(default:'.')) – can be used to indicate a function outside this module, e.g.,my_module.submodulewhich contains the function.
- Return type:
- Returns:
: resulting string
calibration#
Module containing analysis utilities for calibration procedures.
In particular, manipulation of data and calibration points for qubit readout calibration.
- has_calibration_points(s21, indices_state_0=(-2,), indices_state_1=(-1,))[source]#
Determine if dataset with S21 data has calibration points for 0 and 1 states.
Three pieces of information are used to infer the presence of calibration points:
The angle of the calibration points with respect to the average of the datapoints,
The distance between the calibration points, and
The average distance to the line defined be the calibration points.
The detection is made robust by averaging 3 datapoints for each extremity of the “segment” described by the data on the IQ-plane.
See also
- Parameters:
s21 (
ndarray) – Array of complex datapoints corresponding to the experiment on the IQ plane.indices_state_0 (
tuple(default:(-2,))) – Indices in thes21array that correspond to the ground state.indices_state_1 (
tuple(default:(-1,))) – Indices in thes21array that correspond to the first excited state.
- Return type:
- Returns:
: The inferred presence of calibration points.
readout_calibration_analysis#
Module containing an analysis class for two-state readout calibration.
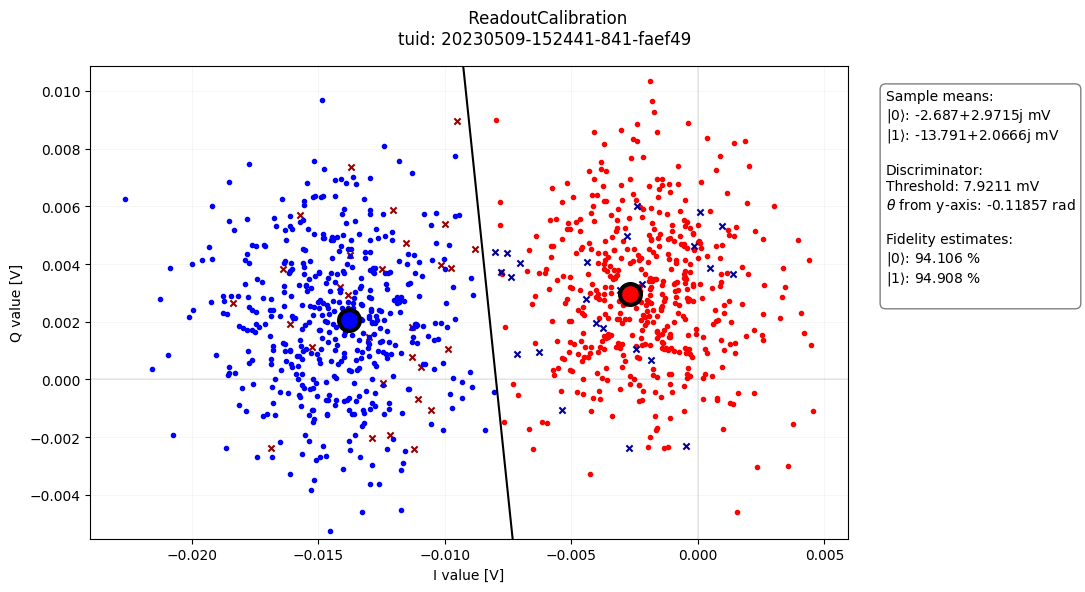
- class ReadoutCalibrationAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisFind threshold and angle which discriminates qubit state.
Example
import os import quantify_core.data.handling as dh from quantify_core.analysis.readout_calibration_analysis import ( ReadoutCalibrationAnalysis, ) # load example data test_data_dir = "../tests/test_data" dh.set_datadir(test_data_dir) ReadoutCalibrationAnalysis(tuid="20230509-152441-841-faef49").run().display_figs_mpl()
- analyze_fit_results()[source]#
Check the fit success and populate
.quantities_of_interest.- Return type:
- create_figures()[source]#
Generate figures of interest.
matplotlib figures and axes objects are added to the
.figs_mpland.axs_mpldictionaries, respectively.- Return type:
conditional_oscillation_analysis#
Module containing an analysis class for the conditional oscillation experiment.
- class ConditionalOscillationAnalysis(dataset=None, tuid=None, label='', settings_overwrite=None, plot_figures=True)[source]#
Bases:
BaseAnalysisAnalysis class for the conditional oscillation experiment.
For a reference to the conditional oscillation experiment, please see section D in the supplemental material of this paper: https://arxiv.org/abs/1903.02492
Example
from quantify_core.analysis.conditional_oscillation_analysis import ( ConditionalOscillationAnalysis ) import quantify_core.data.handling as dh # load example data test_data_dir = "../tests/test_data" dh.set_datadir(test_data_dir) # run analysis and plot results analysis = ( ConditionalOscillationAnalysis(tuid="20230509-165523-132-dcfea7") .run() .display_figs_mpl() )
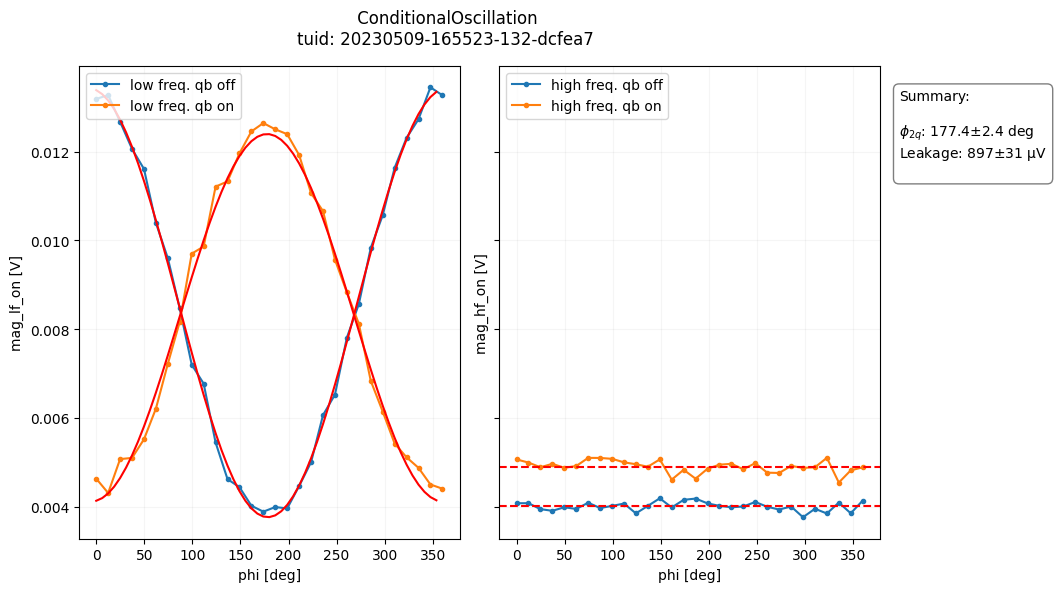
- analyze_fit_results()[source]#
Check fit success and populates
.quantities_of_interest.- Return type:
- create_figures()[source]#
Generate figures of interest.
matplolib figures and axes objects are added to the .figs_mpl and .axs_mpl dictionaries., respectively.
- Return type: